ABSTRACT
Objective: To evaluate the rapid diagnosis of multidrug-resistant tuberculosis, by using a commercial line probe assay for rifampicin and isoniazid detection (LPA-plus), in the routine workflow of a tuberculosis reference laboratory. Methods: The LPA-plus was prospectively evaluated on 341 isolates concurrently submitted to the automated liquid drug susceptibility testing system. Results: Among 303 phenotypically valid results, none was genotypically rifampicin false-susceptible (13/13; 100% sensitivity). Two rifampicin-susceptible isolates harboured rpoB mutations (288/290; 99.3% specificity) which, however, were non-resistance-conferring mutations. LPA-plus missed three isoniazid-resistant isolates (23/26; 88.5% sensitivity) and detected all isoniazid-susceptible isolates (277/277; 100% specificity). Among the 38 (11%) invalid phenotypic results, LPA-plus identified 31 rifampicin- and isoniazid-susceptible isolates, one isoniazid-resistant and six as non-Mycobacterium tuberculosis complex. Conclusions: LPA-plus showed excellent agreement (≥91%) and accuracy (≥99%). Implementing LPA-plus in our setting can speed up the diagnosis of multidrug-resistant tuberculosis, yield a significantly higher number of valid results than phenotypic drug susceptibility testing and provide further information on the drug-resistance level.
Keywords:
Tuberculosis, multidrug-resistant; Molecular diagnostic techniques; Microbial sensitivity tests; Mycobacterium tuberculosis.
RESUMO
Objetivo: Avaliar o diagnóstico rápido de tuberculose multirresistente, utilizando um teste comercial de sondas em linha (LPA-plus), na rotina de um laboratório de referência de tuberculose. Métodos: O teste LPA-plus foi avaliado prospectivamente em 341 isolados simultaneamente submetidos ao teste de suscetibilidade aos antimicrobianos em meio líquido, pelo sistema automatizado. Resultados: Entre os 303 resultados fenotipicamente válidos, nenhum foi genotipicamente falso suscetível à rifampicina (13/13; 100% de sensibilidade). Dois isolados sensíveis à rifampicina apresentavam mutações no gene rpoB (288/290; especificidade de 99,3%), as quais, no entanto, não são associadas à resistência a rifampicina. O LPA-plus não identificou resistência à isoniazida em três isolados fenotipicamente resistentes (23/26; 88,5% de sensibilidade) e detectou todos os isolados sensíveis à isoniazida (277/277; especificidade de 100%). Entre os 38 (11%) resultados fenotípicos inválidos, o LPA-plus identificou 31 isolados sensíveis à rifampicina e à isoniazida, um resistente à isoniazida e seis como micobactérias não tuberculosas. Conclusões: O LPA-plus mostrou excelente concordância (≥91%) e acurácia (≥99%). Sua implementação pode acelerar o diagnóstico da tuberculose multirresistente, produzir número significativamente maior de resultados válidos do que o teste fenotípico de suscetibilidade aos antimicrobianos e fornecer informações adicionais sobre o nível de resistência aos fármacos.
Palavras-chave:
Tuberculose resistente a múltiplos medicamentos; Técnicas de diagnóstico molecular; Testes de sensibilidade microbiana; Mycobacterium tuberculosis.
INTRODUCTIONA major challenge to the effective control of tuberculosis (TB) worldwide is the occurrence of Mycobacterium tuberculosis complex (MTBC) strains showing resistance to both rifampicin (RIF) and isoniazid (INH), the two most effective first-line drugs in TB treatment.(1) This resistance profile, called "multidrug-resistant TB" (MDR-TB), leads to therefore less efficient drug regimens,(1) and is associated with treatment failures, relapses, and poor clinical outcomes.(2)
MDR-TB has called for an urgent development of rapid and accurate diagnostic testing, in order to start effective treatment earlier and reduce the spread of drug-resistant TB.(3,4) To that end, in 2008, the World Health Organization (WHO) endorsed the use of molecular assays for MDR-TB screening.(5) One of them, the GenoType MTBDRplus (Hain Lifescience, Nehren, Germany), is a line-probe assay that detects MTBC, as well as mutations and wild type sequences in the 81-base-pair hotspot region of the rpoB gene, in codon 315 of katG gene, and in the promoter region of inhA gene.(6) MTBDRplus thus predicts MDR-TB by detecting resistance not only to RIF (rpoB gene) but also to INH (katG and inhA genes). Although RIF resistance has been considered a surrogate of MDR-TB,(4,7) identifying INH resistance can be useful, mainly in high TB burden regions in which prevalence of MDR-TB is low,(4) as in Brazil, where 1.5% and 8.0% of the 82,676 TB cases reported in 2016 were estimated as primary and acquired MDR-TB, respectively.(8)
MTBDRplus has shown good accuracy and is now routinely used in many countries,(4) speeding up the MDR-TB diagnosis and reducing the laboratory demand for conventional drug susceptibility testing (DST). However, no studies using this test applied to isolates in the diagnostic workflow of a reference laboratory were conducted so far in Brazil.
We aimed to prospectively evaluate the performance of the MTBDRplus assay applied to MTBC cultures in comparison to phenotypic DST in a high-volume TB reference laboratory, as well as elucidate any discrepancies between the two methods.
METHODSStudy site and populationThis study was conducted in the Tuberculosis and Mycobacteriosis Core of Instituto Adolfo Lutz (IAL), a state governmental institution of São Paulo. The IAL is the TB and mycobacteriosis reference laboratory for São Paulo, responsible for 291 laboratories state wide with different infrastructures for processing the clinical material collected from in- and outpatients, covered by the publicly funded health care system. These laboratories perform acid-fast bacilli smear microscopy or the Xpert MTB/RIF assay (Cepheid, SUNV, CA, USA) implemented in 36 of them by the end of 2014. Eighty laboratories in this network perform cultures and send them to IAL, where DST is performed for patients at higher risk of drug-resistant TB. These include any smear-positive cases after 2 months on TB treatment, those who are contacts of known resistant TB patients, retreatment TB cases, and any immunosuppressed persons, alcohol abusers or illicit drugs users, healthcare workers, homeless individuals, indigenous, immigrants, inmates, inpatients, and prison officers.(9) In 2016, this laboratory framework in São Paulo virtually served a population of 44.85 million inhabitants with a TB incidence rate of 36.4 per 100,000.(10) IAL receives per year approximately 7,000 mycobacterial cultures to confirm identification and performs first-line DST on nearly 4,000 isolates.
For this study, a sample size (n) of 307 TB cases was calculated by using the formula n=Z2P(1-P)/d2(11) applied to an expected 15% (p=0.15) frequency of resistance to at least one of the anti-TB drugs, RIF or INH, and 95% of confidence interval (95%CI), with Z value of 1.96, with 4% precision (d=0.04).
Demographic and clinical data were collected from the Hospital Information and Management System and the TBWeb - Sistema de Controle de Pacientes com Tuberculose (TBWEB) of the state of São Paulo.
Identification of Mycobacterium tuberculosis complex isolatesPrimary mycobacterial cultures referred to IAL in liquid mycobacteria growth indicator tube (MGIT) or on solid media were presumptively identified by observing growth and microscopic characteristics to differentiate MTBC from nontuberculous mycobacteria (NTM). Subsequent identification by phenotypic tests, including MPT64 protein detection, was carried out whenever needed, as already described.(12,13)
Phenotypic drug susceptibility testingPresumptive MTBC isolates were subjected to DST on the automated BACTEC MGIT 960 system (Becton, Dickinson & Co., NJ, EUA),(14) using a modified protocol best suited to the IAL routine conditions.(13) Final concentrations were 0.1 µg INH mL-1 and 1.0 µg RIF mL-1. In case of contamination or absence of growth, the respective primary culture was submitted to further speciation.
Genotype MTBDRplus version 2.0This assay was prospectively performed on 341 isolates, one per patient, that were about to undergo MGIT DST. Cultures underwent DNA extraction on August and October 2014, a day before they entered the MGIT instrument for DST.
DNA extraction from liquid or solid cultures was done using Genolyse kit version 1.0 (Hain)(15) for no more than 23 isolates and a negative control at a time. MDRTBplus was carried out as explained elsewhere,(15) and the reactions detected on strips were visually interpreted with the aid of a cardboard template. In case of invalid results such as no signal with conjugate or any of the other control probes, and doubtful reactions as weak signals with the gene bands, the test was repeated using new DNA extraction.
Gene sequencingSanger sequencing was performed whenever results between MTBDRplus and phenotypic DST remained discordant upon repeating both tests. Isolates showing conflicting results for INH had the mabA-inhA regulatory region (positions -168 to 80, relative to codon) amplified and sequenced with primers mabA-inhAF and mabA-inhAR,(16) as well as the entire inhA and katG genes by using the primer pairs inhA3 and inhA4, inhA3F and inhA5R, and the forward and reverse primers katG-P4, -P5, -P6, -P7 and -P8.(17) For isolates with RIF-discordant results, primers RPOB-1 and RPOB-2(18) were used to amplify and sequence a 350-bp fragment of rpoB encompassing the RIF resistance-determining region.
Single PCR included 12.5 µL of PrimeSTAR Max DNA Polymerase (Takara Bio, Shiga, Japan), 5 pmol of primers for mabA-inhA and katG, 10 pmol of primers for inhA and rpoB, 2 µL of DNA template and PCR-grade water for a final volume of 25 µL. Amplification comprised 30 cycles of 98 °C for 10 seconds, 55 °C for 5 seconds, and 72 °C for 20 seconds. Amplimers purified with ExoSAP-it (Affymetrix, SCL, CA, USA) were sequenced with an ABI 3130xL Genetic Analyzer and the BigDye Terminator version 3.1 Kit (Applied Biosystems, FSTC, CA, USA). Sequences were aligned and analysed using the BioEdit v7.2.5 software(19) and the web-based MUBII-TB-DB(20) and BLAST(21) tools.
Turnaround time of resultsThe time taken to perform MGIT DST and MTBDRplus assays was recorded to calculate the mean time taken to complete the tests. Turnaround time (TAT) of results was calculated from the date oleic acid-albumin-dextrose-catalase (OADC) supplement and antimicrobial solutions were added to MGIT tubes to the date DST result reporting was available; and from the DNA extraction date to the date MTBDRplus result was written on the evaluation sheet.
Data analysesThe sensitivity, specificity, positive (PPV) and negative (NPV) predictive values of the MTBDRplus test were assessed for RIF and INH compared to the phenotypic DST. Agreement between both tests was calculated using kappa (κ) coefficient and the strength of agreement interpreted as poor (< 0.2), fair (> 0.2 ≤ 0.4), moderate (> 0.4 ≤ 0.6), good (> 0.6 ≤ 0.8) and very good (> 0.8 ≤ 1).(22) Two-tailed Fisher's Exact test was used for comparisons between proportions. Differences in TATs were evaluated using paired t test. The significance threshold was set at .05. Statistical analyses were performed using the web-based OpenEpi program.(23)
Ethical StatementThe Technical Scientific Council (CTC-IAL no. 98C/2012) and Research Ethics Committee (CEPIAL no. 207.606 dated Feb-21-2013) of IAL approved this study.
RESULTSPhenotypic drug susceptibility testingThe results of MGIT DST, along with the demographic and clinical characteristics of the patients, are shown in Table 1. Most patients were men (80%), had pulmonary TB (93%) and no past history of TB treatment (65%). MDR-TB was observed only in previously treated pulmonary TB patients.
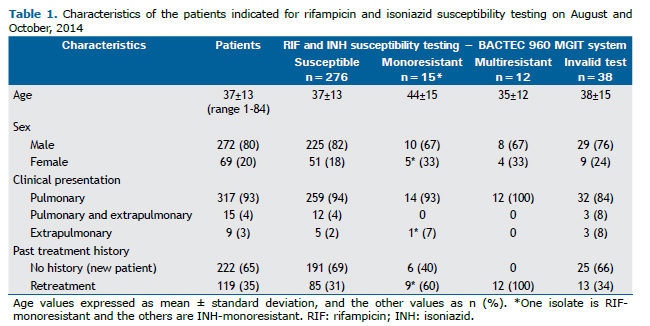
Figure 1 shows the study plan of the 341 isolates prospectively tested. Phenotypic DST provided interpretable results for 303 (89%) isolates, of which 276 (91%) were susceptible, 14 (5%) INH-monoresistant, 12 (4%) MDR, and one (<1%) was RIF-monoresistant.
For the 38 cultures with invalid DST results due to contamination (n=35) or absence of growth (n=3), subsequent speciation identified 23 MTBC, six mixed MTBC + NTM and four NTM cultures. Among the remaining five isolates, identification was not assessed due to insufficient growth of three primary cultures and to heavy contamination in two cases, both Ag MPT64-negative, reported as non-MTBC isolates.
Genotype MTBDRplusAll doubtful (n=9) and invalid (n=2) results became valid upon repeating the assay. MTBDRplus gave interpretable results for all 341 isolates (Figure 1). Among the 335 isolates identified as MTBC, there were 308 (92%) susceptible, 12 (3.6%) MDR, 12 (3.6%) INH-monoresistant and three (1%) RIF-monoresistant isolates.
MTBDRplus presented significantly higher interpretable results, providing information on 38 additional isolates (11%; 95%CI 8.1-14.8%; p<0.0001) for which no MGIT DST results were available (Figure 1). Among these isolates, the genotypic test identified 32 MTBC (31 susceptible and one INH-monoresistant) and six non-MTBC isolates. The one INH-monoresistant and seven susceptible isolates were later confirmed by MGIT DST, on a second isolate.
For RIF resistance prediction, MTBDRplus showed 100% sensitivity (13/13), 99.3% specificity (288/290) and 99.3% accuracy (301/303), as shown in Table 2. The test correctly detected INH resistance in 23/26 isolates (sensitivity 88.5%) and INH susceptibility in all 277 isolates (specificity 100%), with an overall diagnostic accuracy of 99.0% (300/303). PPV and NPV values were high for RIF resistance, INH resistance and MDR, ranging from 86.7% to 100%. The agreement between the genotypic and the phenotypic tests was very good (κ ≥ 0.91). To ascertain the test reproducibility, all the 31 repetitions confirmed the first results.
 Discordances between tests
Discordances between testsWhen the 303 valid results by the phenotypic test were compared to the genotypic test results, there were seven mismatches. After re-growing and re-examining these isolates, two of five initially INH-resistant isolates by the conventional DST matched the MTBDRplus results. The results of the five isolates that remained discrepant are summarized in Table 2. The two false-resistant RIF isolates had their rpoB mutations confirmed as His526Asn and Asp516Tyr by sequencing.
MTBDRplus failed to detect INH resistance in two phenotypically INH-monoresistant and one MDR isolates. Gene sequencing showed the first two isolates had neither katG nor inhA gene mutations while the MDR isolate presented a T nucleotide insertion between positions 1,798 and 1,799 of katG, leading to the Lys600Ile mutation and to a frameshift ending with a stop codon (TGA) at position 623 in the shifted reading frame.
Mutations in rpoB, katG and inhA genesTen different mutation profiles were identified among the 27 genotypically resistant isolates, as shown in Table 3. Regarding the rpoB gene mutations, the most frequent was Ser531Leu (11/15; 73%), mostly among phenotypically MDR isolates (10/12; 83%). His526Asn rpoB mutation alone was observed in one phenotypically susceptible isolate and in the only MDR isolate presenting double mutations in the rpoB gene and concurrent katG and inhA mutations. Ser315Thr1 (AGC→ACC exchange) was the most frequent katG mutation (14/16, 88%) and was harboured mostly by MDR isolates (10/12; 83%). The only mutation found in the inhA gene was C-15T (10; 100%), which was more frequent in INH-monoresistant isolates (8/12; 67%).
Turnaround time of resultsFor the MTBDRplus assay, two consecutive DNA extraction rounds comprising 11 isolates and one control each took about 3 hours. Amplification mix, thermo cycling, hybridization and interpretation of results in one round of 24 samples took 50 minutes, 1 hour and 50 minutes, 2 hours and 20 minutes, and 40 minutes respectively. Therefore, the average TAT from DNA extraction to reporting the results of 24 samples performed by one person alone was 8 to 9 hours overall.
TATs of both MGIT DST and MTBDRplus assays were compared using only valid results on conventional DST. The median TAT to reporting MTBDRplus results was 3 days (zero to 17 days), significantly shorter than that of MGIT DST (median 11 days, 7 to 78 days; p<0.0001). Intervals > 9 days for 16 MTBDRplus results were due to temporary unavailability of the kit, and those > 23 days for 12 MGIT DST reports release were due to repetition of tests presenting growth failure or contamination. As shown in Figure 2, results by MTBDRplus were available much earlier than by MGIT, even though the test was performed by a single operator and in rounds of 24 isolates. By the 7th day, when the first three (1%) MGIT DST results were reported, there were already 231 (76%) MTBDRplus results available. The number of complete tests by the genotypic assay by the 9th day (287; 95%) was attained only on the 14th day by MGIT DST (285; 94%).
 DISCUSSION
DISCUSSIONThis study evaluates the use of the genotype MTBDRplus assay in the workflow of a routine TB laboratory in South-Eastern Brazil, where nearly 4,000 MTBC isolates from patients at high risk of drug-resistant TB in São Paulo undergo MGIT DST per year. The molecular assay was compared to the reference DST on MGIT 960, and discordant results between both methods were resolved by Sanger sequencing. The last IAL's annual reports estimated 91% valid first-line DST results, of which 91% of the isolates were RIF- and INH-susceptible, 4% were MDR, 4% were INH-resistant but RIF-susceptible and < 1% was RIF-resistant but INH-susceptible (data not shown), confirming that the study sample accurately reflected the population of isolates examined each year.
Our findings are in agreement with a review(24) that estimated pooled sensitivities and specificities for resistance prediction to RIF (91.3% and 97.1%) and INH (89.4% and 98.9%). Regarding the data from Brazil using the MTBDRplus in clinical isolates, our findings are comparable with a study(25) in the State of Minas Gerais, showing sensitivities of 93.3% for RIF, 83.3% for MDR or INH resistance detection, and 100% specificity for both drugs. A study on MDR-TB patients in Ribeirão Preto (SP), revealed 100% and 80% sensitivity in detecting RIF and INH resistance, respectively.(26)
In this study, rpoB Ser531Leu and katG Ser315Thr mutations predominated, as in other settings.(25-28) Identifying the specific mutation associated with drug resistance, which is not possible by phenotypic methods, may provide additional information on the category of resistance and guide therapeutic decision, as to the choice of the treatment regimen.(29) In this study, an isolate carrying the Asp516Val rpoB mutation, which was shown to confer resistance to RIF, but not rifabutin,(7,28) illustrates how genetic tests may help clinicians manage TB-resistant cases. Furthermore, the translation of genetic findings into clinical therapy has relevant implications in the use of INH for resistant TB, since this drug was shown to remain effective depending on the INH resistance-conferring mutation. (30) Usual-dose INH is effective when mutations occur solely in the inhA promoter region, and for mutations in katG only, high-dose INH is still an option for most of patients.(30,31) These findings might explain the INH efficacy in the shorter regimen.
Some rpoB mutations detectable only by the absence of reaction with the wild type probes in the MTBDRplus may not be associated to RIF resistance.(7,28,32) This was the case of two phenotypically RIF-susceptible isolates in this study presenting mutations in rpoB codons 513-519 and 526-529, further identified by sequencing as Asp516Tyr and His526Asn, which have been shown not to be associated to RIF resistance.(7,28) As these are not true discordances with the phenotypic test, no false RIF resistance occurred in our study. The results above clearly demonstrate why in cases in which no reaction with the mutation probes occurs, sequencing or phenotypic DST must be performed to better interpret resistance. We did not find in the published literature the rpoB double mutation Phe505Leu and His526Asn seen in an isolate in this study. The MDR profile of that isolate, also harbouring katG Ser315Thr and inhA C-15T mutations, is similar to the one recently described,(33) which presented Phe505Leu and Asp516Tyr, a RIF resistance-conferring double mutation.(32)
Sensitivity for INH resistance detection was lower, as expected, since it can arise from mutations other than those in codon 315 of katG and in the regulatory region of inhA. According to Brossier et al.,(27) MTBDRplus may miss 8% to 21% of INH-resistant isolates. In this study, MTBDRplus missed 2/25 INH-resistant but RIF-susceptible isolates, and 1/12 MDR isolates. The MDR isolate misdiagnosed as RIF-resistant alone would have been submitted to first-line MGIT DST according to the IAL algorithm currently in use for isolates from Xpert-resistant samples. Therefore, the INH resistance of this isolate would be properly identified by MGIT DST. The true INH resistance of the other two false-negative INH results would probably be correctly detected during the follow-up of TB treatment.
Based on the sensitivity and specificity of the MTBDRplus and considering the prevalence estimates of INH resistance among 4,000 isolates received yearly at IAL, we estimated that this test would miss 34 of 292 INH-resistant isolates. On the other hand, MTBDRplus would provide additional information on RIF and INH susceptibilities of 375 MTBC from a total of 446 isolates with invalid results on MGIT DST, yearly.
The shorter TAT to complete the test makes MTBDRplus a more effective method. Most of the laboratory reports would be released before 1% of MGIT DST reports were available. Additionally, the workload on phenotypic DST performance would be drastically reduced, providing time to accommodate more exams. Therefore, not only would presumptive drug-resistant TB patients be given the opportunity to start treatment earlier with the most appropriate regimen, as observed in Ribeirão Preto,(26) but also we could extend access to at least one DST for all patients, as recommended by the WHO. (24) Moreover, MTBDRplus poses a smaller biohazard risk to the laboratory personnel than the conventional DST as it requires less manipulation of live cultures.
To the authors' knowledge, this is the first prospective study in Brazil assessing the usefulness of MTBDRplus in a reference TB laboratory serving the most populous Brazilian state. It provides information for the implementation of this test into the TB diagnostic algorithm in Brazil. However, the study has several limitations. First, the number of resistant isolates was not large enough to draw more sound conclusions on the frequency and pattern of mutations in our setting. Second, we did not measure the MIC of isolates presenting mutations, mainly the one carrying a combination of two rpoB mutations not described in literature. Finally, we did not investigate the presence of mutations in susceptible isolates, as we only sequenced isolates showing conflicting results or not completely identified by MTBDRplus.
In conclusion, the diagnostic accuracy of the MTBDRplus assay was excellent in detecting MTBC resistance to RIF and INH, and MDR. No phenotypically susceptible isolates were misidentified as MDR, nor were any MDR isolates incorrectly predicted as susceptible to both drugs. The advantages of the test, such as reducing the time to diagnosis, being easy to perform and yielding additional results otherwise invalid by the phenotypic DST preclude its disadvantages, notably the false-susceptible INH results. To accurately diagnose clinical resistance, the association of the nature of mutations with the level of phenotypic susceptibility must be carefully evaluated.
REFERENCES1. Centers for Disease Control and Prevention (CDC). Emergence of Mycobacterium tuberculosis with extensive resistance to second-line drugs--worldwide, 2000-2004. MMWR Morb Mortal Wkly Rep. 2006;55(11):301-5.
2. Orenstein EW, Basu S, Shah NS, Andrews JR, Friedland GH, Moll AP, et al. Treatment outcomes among patients with multidrug-resistant tuberculosis: systematic review and meta-analysis. Lancet Infect Dis. 2009;9(3):153-16.
3. Stop TB Partnership and WHO. The global plan to stop TB 2006-2015. Geneva, Switzerland: World Health Organization; 2006.
4. Bai Y, Wang Y, Shao C, Hao Y, Jin Y. GenoType MTBDRplus Assay for Rapid Detection of Multidrug Resistance in Mycobacterium tuberculosis: A Meta-Analysis. PLoS One. 2016;11(3):e0150321.
5. World Health Organization (WHO). Molecular line probe assays for rapid screening of patients at risk of multidrug-resistant tuberculosis (MDR-TB). Policy statement. Geneva, Switzerland: WHO, 2008.
6. Hillemann D, Weizenegger M, Kubica T, Richter E, Niemann S. Use of the genotype MTBDR assay for rapid detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis complex isolates. J Clin Microbiol. 2005;43(8):3699-703.
7. Domínguez J, Boettger EC, Cirillo D, Cobelens F, Eisenach KD, Gagneux S, Hillemann D, Horsburgh R, Molina-Moya B, Niemann S, Tortoli E, Whitelaw A, Lange C; TBNET; RESIST-TB networks. Clinical implications of molecular drug resistance testing for Mycobacterium tuberculosis: a TBNET/RESIST-TB consensus statement. Int J Tuberc Lung Dis. 2016;20(1):24-42.
8. World Health Organization (WHO). Global tuberculosis report 2017. Geneva, Switzerland: WHO, 2017.
9. Brasil. Ministério da Saúde. Secretaria de Vigilância em Saúde. Departamento de Vigilância Epidemiológica. Manual de recomendações para o controle da tuberculose no Brasil [Internet]. Brasília, DF: Ministério da Saúde; 2011 [cited 2019 Feb 28]. Available from: http://bvsms.saude.gov.br/bvs/publicacoes/manual_recomendacoes_controle_tuberculose_brasil.pdf
10. Brasil. Ministério da Saúde. Indicadores prioritários para o monitoramento do Plano Nacional pelo Fim da Tuberculose como Problema de Saúde Pública no Brasil. Boletim Epidemiológico [Internet]. Brasília, DF: Ministério da Saúde; 2017 [cited 2019 Feb 28]. Available from: http://portalarquivos2.saude.gov.br/images/pdf/2017/marco/23/2017-V-48-N-8-Indicadores-priorit--rios-para-o-monitoramento-do-Plano-Nacional-pelo-Fim-da-Tuberculose-como-Problema-de-Sa--de-P--blica-no-Brasil.pdf
11. Naing L, Winn T, Rusli BN. Medical Statistics. Practical issues in calculating the sample size for prevalence studies. Arch Orofacial Sc [Internet]. 2006 [cited 2019 Feb 28];1:9-14. Available from: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.504.2129&rep=rep1&type=pdf
12. Gallo JF, Pinhata JM, Chimara E, Gonçalves MG, Fukasawa LO, Oliveira RS. Performance of an in-house real-time polymerase chain reaction for identification of Mycobacterium tuberculosis isolates in laboratory routine diagnosis from a high burden setting. Mem Inst Oswaldo Cruz. 2016;111(9):545-50.
13. Adami AG, Gallo JF, Pinhata JM, Martins MC, Giampaglia CM, Oliveira RS. Modified protocol for drug susceptibility testing of MGIT cultures of Mycobacterium tuberculosis by the MGIT 960. Diagn Microbiol Infect Dis. 2017;87(2):108-11.
14. Siddiqi SH, Rüsch-Gerdes S. MGIT 960 Procedure Manual; Foundation for innovative new diagnostics (Find) [Internet]. Geneva, Switzerland; 2006 [cited 2019 Feb 28]. Available from: https://www.finddx.org/wp-content/uploads/2016/02/mgit_manual_nov2006.pdf
15. Crudu V, Stratan E, Romancenco E, Allerheiligen V, Hillemann A, Moraru N. First evaluation of an improved assay for molecular genetic detection of tuberculosis as well as rifampin and isoniazid resistances. J Clin Microbiol. 2012;50(4):1264-9.
16. Perdigão J, Macedo R, João I, Fernandes E, Brum L, Portugal I. Multidrug-resistant tuberculosis in Lisbon, Portugal: A molecular epidemiological perspective. Microbial Drug Resistance. 2008;14(2):133-43.
17. Machado D, Perdigão J, Ramos J, Couto I, Portugal I, Ritter C, et al. High-level resistance to isoniazid and ethionamide in multidrug-resistant Mycobacterium tuberculosis of the Lisboa family is associated with inhA double mutations. J Antimicrob Chemother. 2013;68(8):1728-32.
18. Portugal I, Maia S, Moniz-Pereira J. Discrimination of multidrug-resistant Mycobacterium tuberculosis IS6110 fingerprint subclusters by rpoB gene mutation analysis. J Clin Microbiol. 1999;37(9):3022-4.
19. Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser [Internet]. 1999 [cited 2019 Feb 28];41:95-8. Available from: http://brownlab.mbio.ncsu.edu/jwb/papers/1999hall1.pdf
20. Flandrois JP, Lina G, Dumitrescu O. MUBII-TB-DB: a database of mutations associated with antibiotic resistance in Mycobacterium tuberculosis. BMC Bioinformatics. 2014;15:107
21. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403-10.
22. Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics [Internet]. 1977 [cited 2019 Feb 28];33:159-74. Available from: https://www.dentalage.co.uk/wp-content/uploads/2014/09/landis_jr__koch_gg_1977_kappa_and_observer_agreement.pdf
23. Dean AG, Sullivan KM, Soe MM. OpenEpi: Open Source Epidemiologic Statistics for Public Health, Versão. www.OpenEpi.com [Internet]. [cited 2019 Mar 13]. Available from: http://www.openepi.com/Menu/OE_Menu.htm
24. World Health Organization (WHO). The use of molecular line probe assay for the detection of resistance to isoniazid and rifampicin: policy update. WHO/HTM/TB/2016.12. Geneva, Switzerland: WHO, 2016.
25. Dantas NG, Suffys PN, Carvalho WD, Gomes HM, Almeida IN, Figueiredo LJ, et al. Correlation between the BACTEC MGIT 960 culture system with Genotype MTBDRplus and TB-SPRINT in multidrug resistant Mycobacterium tuberculosis clinical isolates from Brazil. Mem Inst Oswaldo Cruz. 2017;112(11):769-74.
26. Feliciano CS, Nascimento MM, Anselmo LM, Pocente RH, Bellissimo-Rodrigues F, Bollela VR. Role of a GenoType MTBDRplus line probe assay in early detection of multidrug-resistant tuberculosis at a Brazilian reference center. Braz J Med Biol Res. 2015;48(8):759-64.
27. Brossier F, Veziris N, Jarlier V, Sougakoff W. Performance of MTBDRplus for detecting high/low levels of Mycobacterium tuberculosis resistance to isoniazid. Int J Tuberc Lung Dis. 2009;13(2):260-5.
28. Berrada ZL, Lin SY, Rodwell TC, Nguyen D, Schecter GF, Pham L, et al. Rifabutin and rifampin resistance levels and associated rpoB mutations in clinical isolates of Mycobacterium tuberculosis complex. Diagn Microbiol Infect Dis. 2016;85(2):177-81.
29. Cabibbe AM, Sotgiu G, Izco S, Migliori GB. Genotypic and phenotypic M. tuberculosis resistance: guiding clinicians to prescribe the correct regimens. Eur Respir J. 2017;50:1702292.
30. Cambau E, Viveiros M, Machado D, Raskine L, Ritter C, Tortoli E, et al. Revisiting susceptibility testing in MDR-TB by a standardized quantitative phenotypic assessment in a European multicenter study. J Antimicrob Chemother. 2015;70(3):686-96
31. Rieder HL, Van Deun A. Rationale for high-dose isoniazid in the treatment of multidrug-resistant tuberculosis. Int J Tuberc Lung Dis. 2017;21(1):123-4.
32. Miotto P, Tessema B, Tagliani E, Chindelevitch L, Starks AM, Emerson C, et al. A standardised method for interpreting the association between mutations and phenotypic drug resistance in Mycobacterium tuberculosis. Eur Respir J. 2017;50(6). pii: 1701354.
33. Appelgren A, Morquin D, Dufour S, Le Moing V, Reynes J, Lotthé A, et al. Investigation of pre-XDR Beijing Mycobacterium tuberculosis transmission to a healthcare worker in France, 2016. J Hosp Infec. 2017;97(4):414-7