ABSTRACT
Obstructive sleep apnea (OSA) has been associated with oxidative stress and various cardiovascular consequences, such as increased cardiovascular disease risk. Quantitative real-time PCR is frequently employed to assess changes in gene expression in experimental models. In this study, we analyzed the effects of chronic intermittent hypoxia (an experimental model of OSA) on housekeeping gene expression in the left cardiac ventricle of rats. Analyses via four different approaches-use of the geNorm, BestKeeper, and NormFinder algorithms; and 2−ΔCt (threshold cycle) data analysis-produced similar results: all genes were found to be suitable for use, glyceraldehyde-3-phosphate dehydrogenase and 18S being classified as the most and the least stable, respectively. The use of more than one housekeeping gene is strongly advised.
Keywords:
Cell hypoxia; Reference standards; Sleep apnea, obstructive; Cardiovascular diseases; Models, animal; Polymerase chain reaction.
RESUMO
A apneia obstrutiva do sono (AOS) tem sido associada ao estresse oxidativo e a várias consequências cardiovasculares, tais como risco aumentado de doença cardiovascular. A PCR quantitativa em tempo real é frequentemente empregada para avaliar alterações na expressão gênica em modelos experimentais. Neste estudo, analisamos os efeitos da hipóxia intermitente crônica (um modelo experimental de AOS) na expressão de genes de referência no ventrículo cardíaco esquerdo de ratos. Análises a partir de quatro abordagens - uso dos algoritmos geNorm, BestKeeper e NormFinder e análise de dados 2−ΔCt (ciclo limiar) - produziram resultados semelhantes: todos os genes mostraram-se adequados para uso, sendo que gliceraldeído-3-fosfato desidrogenase e 18S foram classificados como o mais e o menos estável, respectivamente. A utilização de mais de um gene de referência é altamente recomendada.
Palavras-chave:
Hipóxia celular; Padrões de referência; Apneia do sono tipo obstrutiva; Doenças cardiovasculares; Modelos animais; Reação em cadeia da polimerase.
Obstructive sleep apnea (OSA), which is a sleep-related breathing disorder with a high worldwide prevalence,(1,2) causes various metabolic, oxidative, and sympathetic disturbances, increasing the risk of cardiovascular disease. (3) The most widely used animal model of OSA, particularly when the focus is on its cardiovascular consequences, is the chronic intermittent hypoxia (CIH) model.(3) The CIH model mimics many of the putative pathological effects of OSA, such as increased blood pressure and sympathetic activity.(4)
Real-time PCR is the most commonly used tool to study gene expression of specific targets. In order to obtain relia-ble results in quantitative real-time PCR, it is necessary to use appropriate reference genes, or housekeeping genes (HKGs),(5) as internal controls for gene expression normalization, which is essential to the evaluation of relative gene expression. However, using certain unstable genes as HKGs can strongly compromise data reliability.(6,7) Therefore, each experimental condition should have an HKG selection step in order to obtain good quality data. The three most widely used algorithms for HKG selection are those provided by the computer programs geNorm,(8) BestKeeper,(9) and NormFinder.(10)
To our knowledge, there have been no studies assessing the stability of HKG expression in the left ventricle, or any other cardiac structure, using in vivo models of hypoxia, most such studies having been performed in vitro. There-fore, the objective of the present study was to report the selection of HKGs in the left cardiac ventricle of rats submit-ted to CIH, using the geNorm, BestKeeper, and NormFinder algorithms to analyze the expression stability of five commonly used HKGs. Our results provide important information for HKG selection in future studies employing a CIH model.
Twenty-two adult male Wistar Hannover rats were submitted to CIH (8 h/day) as described previously.(11) All exper-imental procedures were performed in accordance with current ethical and practical guidelines for the use of labora-tory animals.(12) This study was approved by the Animal Research Ethics Committee of the Federal University of São Paulo (Protocol no. 2025/11).
Animals were assigned to one of three experimental groups: control (n = 6); CIH (six weeks of CIH, n = 7); and CIH+N (six weeks of CIH, followed by two weeks of recovery in normoxia, n = 8). The CIH protocol is described in detail elsewhere,(11) as is the rationale behind the use of a two-week recovery period in the CIH+N group.(13)
After six weeks (the end of the CIH protocol) or eight weeks (the end of the CIH+N protocol), animals were eu-thanized by rapid decapitation. Subsequently, the heart was rapidly removed and washed with saline, after which the left ventricle was dissected. All tissues were rapidly dissected on dry ice and stored at −80°C until extraction.
Total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific Inc., Waltham, MA, USA) and treated with DNase. Quantitative real-time PCR was performed with SYBR Green, as described previously.(11) For our HKG eval-uation, we chose five of the most commonly used genes, from different gene pathways: eukaryotic 18S ribosomal RNA (rRNA); beta-actin (ACTB); beta-2-microglobulin (β2M); glyceraldehyde-3-phosphate dehydrogenase (GAPDH); and hypoxanthine guanine phosphoribosyl transferase (HPRT). The primer sequences for all of those HKGs are de-scribed elsewhere.(11)
Gene expression stability was evaluated using the programs geNorm (Gene Quantification: http://download.gene-quantification.info/), NormFinder (Department of Molecular Medicine, Aarhus University Hospital, Aarhus, Denmark: http://moma.dk/normfinder-software), and BestKeeper (Gene Quantification: http://download.gene-quantification.info/). The geNorm program calculates the average expression stability (M-value) for each gene, lower M-values indicating greater stability. (8) The NormFinder program provides a stability value number for each gene, lower stability values indicating less stability. (10) The BestKeeper program calculates a Pearson's correlation coeffi-cient for each gene, values of p closer to 1.0 indicating greater stability. (9) We also analyzed all of the 2−ΔCt (threshold cycle) data by one-way ANOVA.(14) To test for normality and homogeneity, we employed the Kolmogorov-Smirnov test and Levene's test, respectively. The level of statistical significance was set at p < 0.05.
Our evaluation of RNA quality and integrity demonstrated intact 18S and 28S rRNA. Optimization of primer condi-tions and cDNA concentrations was performed and described previously.(11) The 2−ΔCt values, analyzed independent-ly, did not achieve significance (p > 0.05 for all), although 18S expression trended toward significance (p = 0.076 vs. p > 0.4 for the remaining genes; data not shown). These data indicate that, although all five HKG candidates are suitable for use, the expression of 18S demonstrates a tendency to show greater variability between groups.
In our BestKeeper analysis to identify the best HKGs in CIH models, all five candidate genes presented values that were acceptable (p < 0.01). According to the BestKeeper ranking, GAPDH was the best candidate, followed by ACTB. In addition, 18S presented the lowest Pearson's correlation coefficient, indicating that it was the least stable of the five HKG candidates. All BestKeeper Pearson's correlation coefficients are described in Table 1 and depicted in Figure 1A.

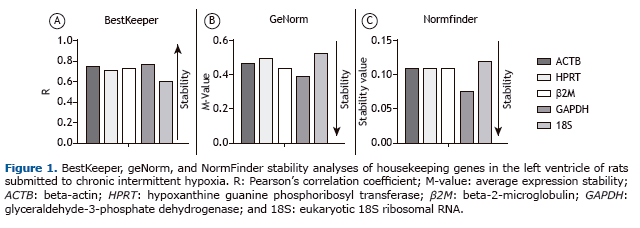
In our geNorm analysis, all five candidate genes presented M-values lower than 1.5, which is considered to be the cut-off value for suitability,(8) in all analyses. According to the geNorm ranking, GAPDH was the best candidate gene, followed by β2M. In addition, 18S presented the highest M-value and was ranked as the least stable gene. All geNorm M-values are described in Table 1 and depicted in Figure 1B. The BestKeeper and geNorm algorithms pro-duced very similar results, the only difference being in terms of the second best HKG candidate.

In our NormFinder analysis, all five candidate genes presented stability values below 0.15, the cut-off value for suitability.(10) Similar although not identical to the BestKeeper results, the NormFinder algorithm identified GAPDH as the best candidate gene, followed by β2M. In addition, 18S presented the highest stability value and was therefore ranked as the least stable gene. All NormFinder stability values are described in Table 1 and depicted in Figure 1C.
The NormFinder HKG ranking was identical to that of BestKeeper, GAPDH and ACTB being ranked as the most sta-ble genes, whereas 18S was ranked as the least stable. However, the geNorm ranking showed a slight difference-ACTB in third position and β2M in second position. The data from all three algorithms corroborate those reported in a previous study involving cardiosphere-derived cells (endogenous cardiac stem cells, candidates for restoring lost tissue) and evaluating the influence of aging on gene stability. The authors found that ACTB, GAPDH, and β2M were the most stable HKGs in both adult or neonatal cardiosphere-derived cells under conditions of normoxia or hypox-ia. (15) Although our study involved the left ventricle portion of the heart, which is composed of several cell types, the tissue origin is the same as that of the cardiosphere-derived cells analyzed by those authors. Conversely, in a study employing an animal model of left ventricular diastolic dysfunction, GAPDH was found to be the second most stable gene, underscoring its stability in vivo.(16) In addition, ACTB presents good stability in models using different hypoxia stimuli, such as various cell types submitted to hypoxia in vitro(7,15) and various brain structures submitted to hypoxia in vivo.(11)
Although analyses of β2M have shown it to be unstable in some brain structures and cell lines,(7,11) our analysis of tissue from the left ventricle ranked β2M among the most stable candidate genes. Our data underscore those of sev-eral other studies, which classified β2M as one of the most stable candidate genes, in cardiosphere-derived cells under hypoxic conditions(15) and in human chondrocytes cultured at oxygen concentrations of 5% and 1%.(17)
In the present study, HPRT was found to be among the least stable candidate genes. However, it can be considered suitable for use because it presented acceptable stability in the geNorm and BestKeeper analyses. Our data are in conflict with those of some previous studies, in which HPRT has been reported to be one of the most stable genes, in cultures of cells submitted to acute hypoxia(7) and in an in vivo model of ischemia in the right ventricle.(18) Neverthe-less, our data corroborate those of another previous study involving cardiosphere-derived cells, in which HPRT was found to be among the least stable genes under conditions of hypoxia.(19) The discrepancies among these studies might be due to differences in the origin of the tissue evaluated (cardiac tissue vs. prostate, kidney, and breast tis-sues) and in the experimental condition (hypoxia vs. ischemia).
Our data corroborate those of several other studies that classified 18S as an unsuitable HKG. In studies employing in vitro models of hypoxia, 18S has been found to be inappropriate as an HKG in the LNCaP (prostate cancer) cell line(20,21) and in locust muscle cells,(6) as it has in studies employing in vivo models of CIH in various brain struc-tures.(11) Our data corroborate those of previous studies demonstrating 18S sensitivity to different hypoxia stimuli. To our knowledge, this is the first study to evaluate 18S expression in cardiac tissue under hypoxic conditions.
Our study has some limitations. The CIH model mimics only one of the four major factors of OSA. In addition, anal-yses of the stability of HKG expression can demonstrate inter-model and even inter-structure variability.(7,11) There-fore, the results of HKG expression studies are not generalizable to all structures or to all models of hypoxia. Never-theless, the results of present study demonstrate that all of the HKGs tested are suitable for use, although it is inad-visable to use 18S. In addition, the geNorm, BestKeeper, and NormFinder algorithms produced very robust results, with little variation among the three. However, to obtain reliable data, the use of more than one HKG is strongly advised.
REFERENCES1. Peppard PE, Young T, Barnet JH, Palta M, Hagen EW, Hla KM. Increased prevalence of sleep-disordered breathing in adults. Am J Epidemiol. 2013;177(9):1006-14. http://dx.doi.org/10.1093/aje/kws342
2. Tufik S, Santos-Silva R, Taddei JA, Bittencourt LR. Obstructive sleep apnea syndrome in the Sao Paulo Epidemiologic Sleep Study. Sleep Med. 2010;11(5):441-6. http://dx.doi.org/10.1016/j.sleep.2009.10.005
3. Dematteis M, Godin-Ribuot D, Arnaud C, Ribuot C, Stanke-Labesque F, Pépin JL, et al. Cardiovascular consequences of sleep-disordered breathing: contribution of animal models to understanding of the human disease. ILAR J. 2009;50(3):262-81. http://dx.doi.org/10.1093/ilar.50.3.262
4. Zoccal DB, Bonagamba LG, Oliveira FR, Antunes-Rodrigues J, Machado BH. Increased sympathetic activity in rats submitted to chronic intermittent hypoxia. Exp Physiol. 2007;92(1):79-85. http://dx.doi.org/10.1113/expphysiol.2006.035501
5. Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, Hennen G, et al. Housekeeping genes as internal standards: use and limits. J Biotechnol. 1999;75(2-3):291-5. http://dx.doi.org/10.1016/S0168-1656(99)00163-7
6. Zhao DJ, Guo K, Kang L. Identification of condition-specific reference genes from microarray data for locusts exposed to hypobaric hypoxia. FEBS Open Bio. 2012;2:235-40. http://dx.doi.org/10.1016/j.fob.2012.08.001
7. Caradec J, Sirab N, Keumeugni C, Moutereau S, Chimingqi M, Matar C, et al. "Desperate house genes": the dramatic example of hypoxia. Br J Cancer. 2010;102(6):1037-43. http://dx.doi.org/10.1038/sj.bjc.6605573
8. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034.
9. Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26(6):509-15. http://dx.doi.org/10.1023/B:BILE.0000019559.84305.47
10. Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245-50. http://dx.doi.org/10.1158/0008-5472.CAN-04-0496
11. Julian GS, de Oliveira RW, Perry JC, Tufik S, Chagas JR. Validation of Housekeeping genes in the brains of rats submitted to chronic intermittent hypoxia, a sleep apnea model. PLoS One. 2014;9(10):e109902. http://dx.doi.org/10.1371/journal.pone.0109902
12. Andersen M, Tufik S. Animal models as ethical tools in biomedical research. São Paulo: Universidade Federal de São Paulo; 2010. 563 pp.
13. Veasey SC, Davis CW, Fenik P, Zhan G, Hsu YJ, Pratico D, et al. Long-term intermittent hypoxia in mice: protracted hypersomnolence with oxidative injury to sleep-wake brain regions. Sleep. 2004;27(2):194-201.
14. Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol. 2006;7:33. http://dx.doi.org/10.1186/1471-2199-7-33
15. Tan SC, Carr CA, Yeoh KK, Schofield CJ, Davies KE, Clarke K. Identification of valid housekeeping genes for quantitative RT-PCR analysis of cardiosphere-derived cells preconditioned under hypoxia or with prolyl-4-hydroxylase inhibitors. Mol Biol Rep. 2012;39(4):4857-67. http://dx.doi.org/10.1007/s11033-011-1281-5
16. Nachar W, Busseuil D, Shi Y, Mihalache-Avram T, Mecteau M, Rhéaume E, et al. Optimisation of reference genes for gene-expression analysis in a rabbit model of left ventricular diastolic dysfunction. PLoS One. 2014;9(2):e89331. http://dx.doi.org/10.1371/journal.pone.0089331
17. Foldager CB, Munir S, Ulrik-Vinther M, Søballe K, Bünger C, Lind M. Validation of suitable house keeping genes for hypoxia-cultured human chondrocytes. BMC Mol Biol. 2009;10:94. http://dx.doi.org/10.1186/1471-2199-10-94
18. Vesentini N, Barsanti C, Martino A, Kusmic C, Ripoli A, Rossi A, et al. Selection of reference genes in different myocardial regions of an in vivo ischemia/reperfusion rat model for normalization of antioxidant gene expression. BMC Res Notes. 2012;5:124. http://dx.doi.org/10.1186/1756-0500-5-124
19. Yao L, Chen X, Tian Y, Lu H, Zhang P, Shi Q, et al. Selection of housekeeping genes for normalization of RT-PCR in hypoxic neural stem cells of rat in vitro. Mol Biol Rep. 2012;39(1):569-76. http://dx.doi.org/10.1007/s11033-011-0772-8
20. Caradec J, Sirab N, Keumeugni C, Revaud D, Loric S. Reply: 18S is an appropriate housekeeping gene for in vitro hypoxia experiments. Br J Cancer. 2010;103(4):591-2. http://dx.doi.org/10.1038/sj.bjc.6605755
21. Nagelkerke A, Mujcic H, Wouters B, Span PN. 18S is an appropriate housekeeping gene for in vitro hypoxia experiments. Br J Cancer. 2010;103(4):590; author reply 591-2. http://dx.doi.org/10.1038/sj.bjc.6605754