ABSTRACT
Objective: Silicosis is a pneumoconiosis characterized by fibrosis of the lung parenchyma caused by inhalation of silica particles. Genetic factors might play a role in the severity silicosis. We sought to evaluate the influence of polymorphisms in the ACE, FAS, FASLG, NOS2, IL1RN, FAM13A, TGFB1, and TNF genes on the severity of silicosis. Methods: Nine polymorphisms were genotyped by PCR in a sample of 143 patients with silicosis in the state of Rio de Janeiro, Brazil. Results: Fifty-seven patients (40%) were classified as having simple silicosis and 86 (60%) were classified as having complicated silicosis. The TT genotype of rs1800469 in the TGFB1 gene showed a protective effect for complicated silicosis (OR = 0.35; 95% CI, 0.14-0.92; p = 0.028) when compared with the other two genotypes (CC+CT). The polymorphic T allele of rs763110 in the FASLG gene (OR = 0.56; 95% CI, 0.31-0.99; p = 0.047), as well as a dominant model for the T allele (TT+CT: OR = 0.37; 95% CI, 0.15-0.96; p = 0.037), also showed a protective effect. When patients with simple silicosis despite having been exposed to silica for a longer time (> 44,229 hours) were compared with patients with complicated silicosis despite having been exposed to silica for a shorter time, the T allele of rs763110 in the FASLG gene (OR = 0.20; 95% CI, 0.08-0.48; p < 0.0001), as well as dominant and recessive models (OR = 0.06; 95% CI, 0.00-0.49; p = 0.01 and OR = 0.22; 95% CI, 0.06-0.77; p = 0.014, respectively), showed a protective effect against the severity of silicosis. Conclusions: It appears that rs1800469 polymorphisms in the TGFB1 gene and rs763110 polymorphisms in the FASLG gene are involved in the severity of silicosis. Given the lack of studies relating genetic polymorphisms to the severity of silicosis, these results should be replicated in other populations.
Keywords:
Silicosis; Polymorphism, genetic; Genetic association studies; Cytokines.
RESUMO
Objetivo: A silicose é uma pneumoconiose caracterizada por fibrose do parênquima pulmonar causada por inalação de partículas de sílica. Fatores genéticos podem desempenhar um papel na gravidade da silicose. Nosso objetivo foi avaliar a influência de polimorfismos dos genes ACE, FAS, FASLG, NOS2, IL1RN, FAM13A, TGFB1 e TNF na gravi-dade da silicose. Métodos: Nove polimorfismos foram genotipados por meio de PCR em uma amostra composta por 143 pacientes com silicose no estado do Rio de Janeiro, Brasil. Resultados: A silicose foi classificada em simples em 57 (40%) dos pacientes e em complicada, em 86 (60%). O genótipo TT do polimorfismo rs1800469 do gene TGFB1 teve efeito protetor contra a silicose complicada (OR = 0,35; IC95%: 0,14-0,92; p = 0,028) em comparação com os outros dois genótipos (CC+CT). O alelo T polimórfico do polimorfismo rs763110 do gene FASLG (OR = 0,56; IC95%: 0,31-0,99; p = 0,047) e um modelo dominante do alelo T (TT+CT: OR = 0,37; IC95%: 0,15-0,96; p = 0,037) tam-bém tiveram efeito protetor. Quando se compararam os pacientes que tinham silicose simples com um tempo maior de exposição à sílica (> 44.229 horas) àqueles que tinham silicose complicada com um tempo menor de ex-posição à sílica, o alelo T do polimorfismo rs763110 do gene FASLG (OR = 0,20; IC95%: 0,08-0,48; p < 0,0001) e mode-los dominantes e recessivos (OR = 0,06; IC95%: 0,00-0,49; p = 0,01 e OR = 0,22; IC95%: 0,06-0,77; p = 0,014, respecti-vamente) tiveram efeito protetor contra a gravidade da silicose. Conclusões: Polimorfismos rs1800469 do gene TGFB1 e polimorfismos rs763110 do gene FASLG parecem estar envolvidos na gravidade da silicose. Como há pou-cos estudos que tenham estabelecido relações entre polimorfismos genéticos e a gravidade da silicose, esses re-sultados devem ser replicados em outras populações.
Palavras-chave:
Silicose; Polimorfismo genético; Estudos de associação genética; Citocinas.
INTRODUCTION Silicosis is the most prevalent pneumoconiosis in the world, being characterized by fibrosis of the lung parenchyma caused by inhalation of silica particles. (1) Silicosis is an occupational disease that can affect workers in the mineral extraction industry, mineral processing industry, manufacturing industry, cosmetics industry, and sandblasting industry, among others.(2,3) The development of silicosis is related to the duration and degree of exposure to silica particles; deficiencies in the immune systems; impaired pulmonary clearance; smoking; the concentration of silica particles; personal protective equipment (PPE) use (or lack thereof); and genetic factors.(4,5) Genetic factors might explain distinct phenotypic expressions of the disease in patients with a similar exposure history.(4,5)
In the alveolar space, silica particles trigger various inflammatory mechanisms and pathways responsible for the pathogenesis of silicosis, represented by cycles of injury and healing. These cycles generate intense epithelial damage causing severe impairment of gas exchange by deposition of collagen fibers in the alveolar space, being divided into the following stages: direct cytotoxicity; generation of reactive oxygen species and reactive nitrogen species; intense production of cytokines; deposition of collagen fibers; fibrosis; apoptosis of alveolar cells; and release of crystals in the alveolar space, restarting the cycle.(1)
Silica particles lead to intensive production of reactive oxygen species and reactive nitrogen species, mainly by alveolar epithelial cells and macrophages. Inducible nitric oxide synthase activity is fundamental to this oxidative environment because it acts in the production of nitric oxide.(6) This intense oxidative environment promotes the release of several interleukins. IL-1 (IL-1α and IL-1β) acts in the activation of fibroblasts and in the deposition of collagen fibers in the alveolar space. However, IL-1 receptor antagonist (IL1RA) acts by blocking the action of IL-1.(7) TNF-α acts in fibroblast recruitment and proliferation, and as a ligand for apoptosis receptors.(1) TGFB1 acts in cell proliferation, differentiation, migration, inflammation, and apoptosis, as well as in tissue repair.(8) Angiotensin-converting enzyme (ACE), released by several epithelial cells and by macrophages, is an important biomarker of lung injury.(9) Family with sequence similarity 13 member A (FAM13A) is expressed in the airways in type II epithelial cells and alveolar macrophages, and seems to play a role in the pathogenesis of idiopathic pulmonary fibrosis.(10)
Alveolar macrophage apoptosis also contributes to the pathogenesis of silicosis, through increased expression of Fas cell surface death receptor (FAS) and its ligands (Fas ligand [FASLG] and TNF-α). Alveolar macrophage apoptosis releases inflammatory mediators, promoting new recruitment of inflammatory cells and repeated activation of inflammatory pathways.(1)
Silicosis is an irreversible and incurable disease. The identification of genetic polymorphisms is essential for early identification of patients who are more likely to present with increased disease severity, making it possible to establish appropriate follow-up strategies and decide on the use of antifibrotic drugs.(11) The objective of the present study was to evaluate the influence of polymorphisms in the ACE (rs4646994), FAS (rs2234767), FASLG (rs763110), nitric oxide synthase 2—NOS2—(rs2297518), IL1RN (rs419598 and rs2234663), FAM13A (rs2609255), TGFB1 (rs1800469), and TNF (rs1800629) genes on the severity of silicosis in patients in Brazil.
METHODS Patients The study sample consisted of 143 patients diagnosed with silicosis on the basis of an occupational history of exposure to silica particles and radiological findings consistent with silicosis, in accordance with the International Labour Organization (ILO) International Classification of Radiographs of Pneumoconioses.(12) Patients with occupational lung diseases other than silicosis were excluded.
The patients included in the study were treated at the Fluminense Federal University Antônio Pedro University Hospital, the State University of Rio de Janeiro Pedro Ernesto University Hospital, or the Oswaldo Cruz Foundation School of Public Health, all of which are located in the state of Rio de Janeiro, Brazil. All patients were removed from work due to confirmed silicosis.
On the basis of chest X-rays findings, patients were classified as having simple silicosis (or small opacities, i.e., opacities smaller than 1.0 cm) or complicated silicosis (or large opacities, i.e., at least one opacity greater than 1.0 cm), in accordance with the ILO International Classification of Radiographs of Pneumoconioses.(12) The radiographs were obtained with the use of an LX30 X-ray machine (Siemens AG, Erlangen, Germany) and were evaluated separately by three ILO-certified readers. All of the study participants gave written informed consent, and the study was approved by the local research ethics committees.
Sociodemographic characteristics, clinical characteristics, and lung function parameters Sociodemographic and clinical characteristics such as age (in years), occupation, weight (in kg), height (in m), BMI (in kg/m2), total number of years of silica exposure, total number of hours worked per week, total number of hours of silica exposure, time elapsed since removal of exposure (in years), smoking history (in pack-years), use of PPE, and tuberculosis were assessed by means of a questionnaire.
Lung function parameters were obtained by spirometry, which was performed with an MS-PFT spirometer (Jaeger, Würzburg, Germany) and in accordance with the American Thoracic Society/European Respiratory Society standards(13) and the Brazilian Thoracic Association standardized methods. (14,15) FEV1, FVC, and the FEV1/FVC ratio were evaluated in all patients.
Genotyping For genetic material collection, cell samples were obtained from the oral cavity by rinsing with 5 mL of saline solution (0.5% NaCl) for 60 s. The samples were then identified and immediately stored in a freezer at −4°C. Genomic DNA extraction was performed in accordance with Aidar & Line.(16)
Genotyping was performed by PCR. Standard PCR was used in order to analyze 2018T/C (rs419598) and 86 bp variable number tandem repeats (rs2234663) in the IL1RN gene, as well as Ins/Del (rs4646994) in the ACE gene. PCR conditions and primers were as previously described.(9,17,18) For ACE rs4646994, the alleles consisted of a 190-bp fragment (D allele) and a 490-bp fragment (I allele), detected by electrophoresis of PCR products on 2% agarose gel. For rs2234663 in the IL1RN gene, genotypes were established according to the sizes of PCR products on 2.5% agarose gel: IL1RN*1 410 bp (four repeats); IL1RN*2 240 bp (two repeats); IL1RN*3 500 bp (five repeats); IL1RN*4 325 bp (three repeats); and IL1RN*5 595 bp (six repeats).
Genotyping of the rs419598 polymorphism was performed by PCR, followed by RFLP with 1 U of MspI, in accordance with the manufacturer instructions (New England Biolabs, Ipswich, MA, USA). Electrophoresis on 3% agarose gel showed three PCR fragment sizes: 123 bp and 233 bp (C allele), and 356 bp (T allele).
Genotyping of FAM13A rs2609255, FAS rs2234767 (−1377G/A), FASLG rs763110 (−844C/T), NOS2 rs2297518 (Ser608Leu), TGFB1 rs1800469 (−509C/T), and TNF rs1800629 (−308G/A) was performed by real-time PCR with predesigned and validated TaqMan® assays (C_15906608_10, C_12123966_10, C_3175437_10, C_11889257_10, C_8708473_10, and C_7514879_10, respectively; Thermo Fisher Scientific, Waltham, MA, USA). The genotyping protocol followed the manufacturer instructions (Thermo Fisher Scientific), and the samples were run in a CFX96 real-time PCR system (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Statistical analysis Allele frequencies were obtained by gene counting. Deviations from the Hardy-Weinberg equilibrium were evaluated by the chi-square test. Association analyses between gene polymorphisms and silicosis severity (simple vs. complicated) were evaluated by the chi-square test or Fisher’s exact test. Quantitative data with normal distribution (as determined by the Kolmogorov-Smirnov test) were evaluated by a t-test, whereas quantitative data with non-normal distribution were evaluated by the Mann-Whitney test. Values were presented as mean ± standard deviation. The total number of hours of silica exposure was obtained by adjusting the total number of hours worked per week in each month (11 months of work) and multiplying it by the total number of years worked. The time elapsed since removal of exposure (in years) was calculated by the difference between the year of silicosis diagnosis (and sick leave) and the year of sample collection and classification as simple silicosis or complicated silicosis.
A multivariate logistic regression analysis was used in order to assess the ORs and 95% CIs for independent predictors of complicated silicosis. The logistic regression model included the following independent variables: polymorphisms significantly associated with silicosis severity, total number of hours of silica exposure > 44,229, time elapsed since removal of exposure (in years), and no use of PPE. The level of significance was set at p < 0.05. All statistical tests were performed with the IBM SPSS software package, version 20.0 (IBM Corporation, Armonk, NY, USA).
RESULTS Sociodemographic characteristics, clinical characteristics, and lung function parameters Of the 143 patients, 57 (40%) were classified as having simple silicosis and 86 (60%) were classified as having complicated silicosis. Sandblasting was the most common occupation, in 66 patients (46%), followed by marble work, in 21 (14.7%), and hammering, in 18 (12.6%). Seventy-six patients (53%) had a history of smoking. Of those, 27 (35.5%) had simple silicosis and 49 (64.5%) had complicated silicosis (p = 0.260). With regard to tuberculosis, 70 patients (49%) had a history of tuberculosis. Of those, 25 (35.7%) had simple silicosis and 45 (64.3%) had complicated silicosis (p = 0.321). With regard to the use of PPE, 54 (37.8%) reported not using PPE at work. Of those, 20 (37%) had simple silicosis and 34 (63%) had complicated silicosis (p = 0.101). Other clinical and demographic characteristics are presented in Table 1.
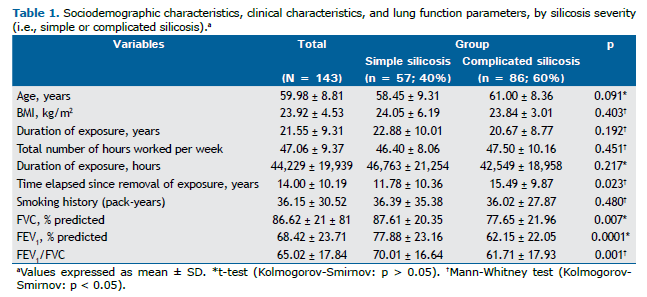
A statistically significant difference was observed between patients with simple silicosis and those with complicated silicosis regarding the time elapsed since removal of exposure (p = 0.023). With regard to lung function parameters, a statistically significant difference was observed between patients with simple silicosis and those with complicated silicosis regarding percent predicted FVC (p = 0.007), FEV1 (p = 0.0001), and FEV1/FVC (p = 0.001; Table 1).
Association analysis between gene polymorphisms and silicosis severity Genotype frequencies of all polymorphisms in the total sample showed no significant deviation from the Hardy-Weinberg equilibrium. Associations between the severity of silicosis (simple silicosis vs. complicated silicosis) and the genetic polymorphisms investigated in the present study were significantly different for −844C>T (rs763110) in the FASLG gene and −509C>T (rs1800469) in the TGFB1 gene (Table 2). The polymorphic T allele of rs763110 in the FASLG gene showed a protective effect for complicated silicosis (OR = 0.56; 95% CI, 0.31-0.99; p = 0.047). In a dominant model, carriers of the T allele (TT+CT) also showed this protective effect (OR = 0.37; 95% CI, 0.15-0.96; p = 0.037; Table 2).
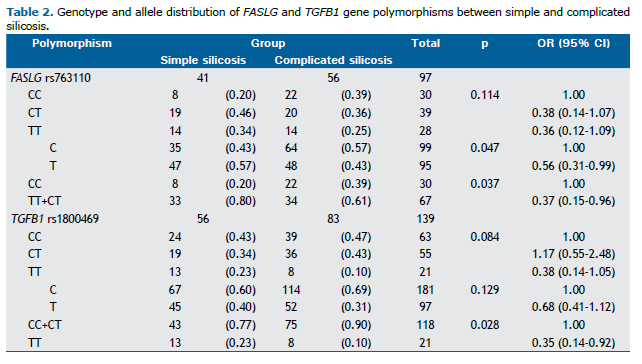
The −509C>T (rs1800469) polymorphism in the TGFB1 gene showed a significant protective effect in a recessive model of the T allele. As can be seen in Table 2, the TT genotype was more common than the other two genotypes (CC+CT) in patients with simple silicosis (OR = 0.35; 95% CI, 0.14-0.92; p = 0.028). For the remaining polymorphisms, no significant associations were observed (Table 2).
No significant differences were observed between patients with simple silicosis and those with complicated silicosis regarding the other polymorphisms: ACE rs4646994 (p = 0.171), FAS rs2234767 (p = 0.709), FAM13A rs2609255 (p = 0.402), IL1RN rs419598 (p = 0.804), IL1RN rs2234663 (p = 0.978), NOS2 rs2297518 (p = 0,221), and TNF-α rs1800629 (p = 0.289; data not shown).
Association analysis between gene polymorphisms and silicosis severity, based on the total number of hours of silica exposure Using the mean number of hours of silica exposure in the sample as a whole (44,229 hours), we compared genotype and allele distributions between simple silicosis patients with > 44,229 hours of exposure to silica and complicated silicosis patients with < 44,229 hours of exposure to silica. Our objective was to compare patients with simple silicosis despite longer exposure to silica and those with complicated silicosis despite shorter exposure to silica. We found a significant association between the two groups of patients and the rs763110 polymorphism in the FASLG gene.
The T allele showed a protective effect against complicated silicosis with increased exposure to silica (OR = 0.20; 95% CI, 0.08-0.48; p < 0.0001). This was also observed in dominant and recessive models of the T allele (OR = 0.06; 95% CI, 0.00-0.49; p = 0.01 and OR = 0.22; 95% CI, 0.06-0.77; p = 0.014, respectively; Table 3).
Multivariate logistic regression analysis Table 4 shows the results of the multivariate logistic regression analysis of independent predictors of silicosis severity. FASLG TT+TC genotypes were found to be predictors of protection against complicated silicosis after controlling for risk factors (OR = 0.15; 95% CI, 0.03-0.75; p = 0.021), whereas no effect was observed for TGFB1 TT genotype. No use of PPE and a longer time elapsed since removal of exposure were also important independent risk factors for complicated silicosis (OR = 6.40; 95% CI, 1.47-27.86; p = 0.013 and OR = 1.09; 95% CI, 1.01-1.17; p = 0.027, respectively).
DISCUSSION A history of exposure to silica particles is the most important of all environmental factors related to silicosis.(19) However, phenotypic differences could be explained by individual responses to exposure, and genetic variations could influence these responses. (4,19) In this study, we evaluated the association of several polymorphisms with the severity of silicosis in workers exposed to silica in Brazil and found significant associations between disease severity and the TGFB1 and FASLG genes.
TGFB1 is a multifunctional cytokine that regulates the proliferation and differentiation of various cell types and is directly involved in fibrosis of the lung parenchyma. This cytokine can bind to at least three receptors (types I, II, and III). The effects of TGFB1 on extracellular matrix synthesis and deposition are mediated by type I receptors; the effects of TGFB1 on cell growth and proliferation are mediated by type II receptors; and type III receptors inhibit the binding of TGFB1 to cell membrane receptors, inhibiting its action.(20) Fibrosis represents a pathological event of a normal tissue repair process. Therefore, excessive or sustained production of TGFB1 becomes a key point for tissue fibrosis. In animal and human models, limited tissue injury is accompanied by a transient increase in TGFB1, without progression to fibrosis. In the presence of repetitive injury, TGFB1 production is maintained, leading to progressive extracellular matrix deposition and fibrosis.(20) The mechanism involved in the maintenance of TGFB1 expression as a result of repetitive injury is still poorly understood.(20)
The rs1800469 (−509C/T) promoter polymorphism in the TGFB1 gene alters the levels of TGFB1 production and secretion.(21) Plasma levels of TGFB1 are twice as high in patients homozygous for the T allele in comparison with individuals homozygous for the C allele, with heterozygotes showing intermediate production.(21,22) In a meta-analysis performed by Deng et al. and including seven studies (a total of 4,333 patients with pneumoconiosis and 3,478 controls), a significant association was found between the rs1800469 polymorphism and the risk of pneumoconiosis.(8) Wu et al. conducted an association study of the rs1800469 polymorphism in 183 patients with silicosis and 111 controls and observed no significant association. (21) In our study, we observed that the TT genotype represented a protective factor for silicosis severity.
The divergence in results might be due to the fact that TGFB1 also acts as an anti-inflammatory agent in silicosis, playing an important role in the initiation and termination of tissue repair after an aggression, leading to tissue remodeling.(20) This was confirmed by Barbarin et al.,(23) who demonstrated that the pathogenesis of silicosis involves a complex interaction of inflammatory and anti-inflammatory pathways. TGFB1 stimulates fibroblasts and smooth muscle cells to produce elastin, as well as stimulating the production of inflammatory mediators.(24) TGFB1 also acts as an anti-inflammatory agent, promoting extracellular matrix accumulation by decreasing collagenase synthesis; repressing the stimulatory effects of growth factors on collagenase gene expression; and increasing the production of collagenase inhibitors.(24) Moreover, TGFB1 acts in all stages of tissue repair, inhibiting T and B cells and their products (TNF-α and IL-1), as well as modulating macrophage cytotoxicity, including suppression of superoxide and nitric oxide production.(25)
Experimental studies have demonstrated that administration of TGFB1 results in normalization of the tissue damage process.(26) In addition, the different expressions of its receptors (I, II, and III) and interactions with TGFB1 may cause an imbalance between the inflammatory and anti-inflammatory pathways. There are no studies evaluating the association of TGFBR1 polymorphisms with silicosis or other pneumoconioses. There are also no studies evaluating the association of TGFB1 and TGFBR1 polymorphisms with silicosis or other pneumoconioses. However, some studies have investigated other diseases. Grigorova et al.(27) evaluated the association of the rs1800469 polymorphism in the TGFB1 gene and the rs3087465 polymorphism in the TGFBR2 gene with frequent episodes of disease activity in patients with multiple sclerosis. The authors found a higher concentration of TGFB1 determined by the TGFB1 genotype in combination with the TGFBR2 genotype and concluded that this combination acted as a protective factor for relapsing-remitting multiple sclerosis.(27) Jin et al.(28) analyzed the association of the aforementioned polymorphisms with esophageal squamous cell carcinoma. The authors concluded that individuals carrying variant genotypes of these polymorphisms had a significantly reduced risk of esophageal squamous cell carcinoma.
Apoptosis plays an important role in the pathophysiology of silicosis.(29) The FAS receptor, located at 10q24.1, is a potent member of the apoptosis receptor family, playing a key role in signaling for apoptosis of several cells. This receptor interacts with its ligand (FASLG), initiating the cell death cascade by apoptosis.(30) In the FAS/FASLG system, the FAS receptor is expressed in several cells of various tissues, whereas FASLG is restricted to cells of the immune system, such as activated T cells and natural killer cells.(31) Borges et al.(32) investigated the role of FASLG in silicosis and found that FASLG-deficient guinea pigs were resistant to silicosis because of reduced macrophage apoptosis. In addition, treatment with an FASLG antagonist antibody prevented the development of the disease.(32)
In the present study, the T allele of the rs763110 polymorphism in the FASLG gene showed a protective effect on disease severity, even when we compared patients with simple silicosis and longer exposure to silica with those with complicated silicosis and shorter exposure to silica. Studies have shown that the T allele of this polymorphism leads to lower FASLG expression(33) because the TT genotype also leads to suppression of apoptosis by reduced binding of the FASLG promoter to transcription factors.(34) Cooke et al. demonstrated that the C allele of this polymorphism causes higher basal expression of FASLG than does the T allele.(35) Wu et al.(4) analyzed the influence of this polymorphism on susceptibility to silicosis and found no significant association. In contrast, in our study, the protective effect of rs763110 in FASLG for the severity of silicosis might be due to reduced apoptosis caused by the presence of the T allele in patients with simple silicosis.
Our multivariate logistic regression analysis of independent predictors of silicosis severity revealed that FASLG TT+TC genotypes constituted an independent protection factor for complicated silicosis (OR = 0.15; 95% CI, 0.03-0.75; p = 0.021). It also revealed that no use of PPE (OR = 6.40; 95% CI, 1.47-27.86; p = 0.013) was an independent risk factor for complicated silicosis. Recently, Requena-Mullor et al. studied silicosis in artificial stone workers and demonstrated that using only a simple mask and not using the PPE provided by the company represented a greater risk for silicosis.(36) Our multivariate analysis also revealed that a longer time elapsed since removal of exposure was also an independent risk factor for complicated silicosis (OR = 1.09; 95% CI, 1.01-1.17; p = 0.027). In silicosis, fibrosis can progress as a result of the inflammatory process caused by silica dust, even after removal of exposure. Therefore, silicosis tends to progress even in patients who have been away from occupational exposure for a long time.
The present study has limitations, including the impossibility of analyzing all of the pathogenic pathways of silicosis, including IL receptors; the presence of memory bias (the exact duration of exposure and number of hours worked per week); appropriate PPE and its use; and the small sample size. Another limitation is that we analyzed chest X-rays and classified the findings on the basis of the ILO International Classification of Radiographs of Pneumoconioses.(12) However, several studies have shown greater sensitivity with the use of chest CT scans in the assessment of profusion of lung parenchymal opacities in patients with silicosis.(37-39) Yet another limitation is related to the inclusion of patients with tuberculosis, which is known to promote progression of parenchymal lesions caused by silicosis. However, the exclusion of these patients would have resulted in a small sample size and would have misrepresented the profile of silicosis patients in Brazil.
In conclusion, the pathophysiology of silicosis is extremely complex, encompassing inflammatory and anti-inflammatory pathways. Polymorphisms in genes related to several immune mechanisms could cause an imbalance between inflammatory and anti-inflammatory factors, explaining the expression of distinct phenotypes. This study identified a statistically significant difference in the prevalence of TGFB1 and FASLG gene polymorphisms, which were found to act as protective factors for disease severity. Further studies should be carried out in an attempt to expand the sample size and analyze the associations between genetic polymorphisms and pathophysiological mechanisms.
ACKNOWLEDGMENTS We wish to thank all of the patients who volunteered to participate in this study, as well as their relatives.
CONFLICTS OF INTEREST None declared.
AUTHOR CONTRIBUTIONS FBK and ASFN: study concept and design; MCSC, ASFN, KCRS, HC, PC, WC, and FBK: sample collection; MCSC, KCRS, JFBS, CBMN, and FBK: data collection, analysis, and interpretation; MCSC, JMR, and FBK: drafting of the manuscript and critical revision of the manuscript for important intellectual content; MCSC, ASFN, KCRS, JFBS, JMR, HC, PC, WC, CBMN, and FBK: final approval of the submitted version.
REFERENCES 1. Lopes-Pacheco M, Bandeira E, Morales MM. Cell-Based Therapy for Silicosis. Stem Cells Int. 2016;2016:5091838. https://doi.org/10.1155/2016/5091838
2. Hoy RF, Baird T, Hammerschlag G, Hart D, Johnson AR, King P, et al. Artificial stone-associated silicosis: a rapidly emerging occupational lung disease. Occup Environ Med. 2018;75(1):3-5. https://doi.org/10.1136/oemed-2017-104428
3. The Lancet Respiratory Medicine. The world is failing on silicosis. Lancet Respir Med. 2019;7(4):283. https://doi.org/10.1016/S2213-2600(19)30078-5
4. Wu F, Xia Z, Qu Y, Tang Y, Cao D, Sun P, Christiani DC. Genetic polymorphisms of IL-1A, IL-1B, IL-1RN, NFKB1, FAS, and FASL, and risk of silicosis in a Chinese occupational population. Am J Ind Med. 2008;51(11):843-851. https://doi.org/10.1002/ajim.20616
5. Fernández Álvarez R, Martínez González C, Quero Martínez A, Blanco Pérez JJ, Carazo Fernández L, Prieto Fernández A. Guidelines for the diagnosis and monitoring of silicosis. Arch Bronconeumol. 2015;51(2):86-93. https://doi.org/10.1016/j.arbres.2014.07.010
6. Qu Y, Tang Y, Cao D, Wu F, Liu J, Lu G, et al. Genetic polymorphisms in alveolar macrophage response-related genes, and risk of silicosis and pulmonary tuberculosis in Chinese iron miners. Int J Hyg Environ Health. 2007;210(6):679-689. https://doi.org/10.1016/j.ijheh.2006.11.010
7. Zhou Y, Kang Y, Zhang Z, Liu J. IL-1RA +2018 polymorphism and the susceptivity to pneumoconiosis: a Meta-analysis. Int J Clin Exp Med. 2014;7(8):2204-2208.
8. Deng CW, Zhang XX, Lin JH, Huang LF, Qu YL, Bai C. Association between Genetic Variants of Transforming Growth Factor-β1 and Susceptibility of Pneumoconiosis: A Meta-analysis. Chin Med J (Engl). 2017;130(3):357-364. https://doi.org/10.4103/0366-6999.198917
9. Mohebbi I, Abdi Rad I, Bagheri M. Association of angiotensin-1-converting enzyme gene variations with silicosis predisposition. Inhal Toxicol. 2010;22(13):1110-1115. https://doi.org/10.3109/08958378.2010.526654
10. Hirano C, Ohshimo S, Horimasu Y, Iwamoto H, Fujitaka K, Hamada H, et al. FAM13A polymorphism as a prognostic factor in patients with idiopathic pulmonary fibrosis. Respir Med. 2017;123:105-109. https://doi.org/10.1016/j.rmed.2016.12.007
11. Flaherty KR, Wells AU, Cottin V, Devaraj A, Walsh SLF, Inoue Y, et al. Nintedanib in Progressive Fibrosing Interstitial Lung Diseases. N Engl J Med. 2019;381(18):1718-1727. https://doi.org/10.1056/NEJMoa1908681
12. International Labour Organization (ILO). Guidelines for the use of ILO International classification of radiographs of pneumoconioses, rev ed. Occupational Safety and Health Series n..22. Geneva: ILO; 2011.
13. Graham BL, Steenbruggen I, Miller MR, Barjaktarevic IZ, Cooper BG, Hall GL, et al. Standardization of Spirometry 2019 Update. An Official American Thoracic Society and European Respiratory Society Technical Statement. Am J Respir Crit Care Med. 2019;200(8):e70-e88. https://doi.org/10.1164/rccm.201908-1590ST
14. Neder JA, Andreoni S, Castelo-Filho A, Nery LE. Reference values for lung function tests. I. Static volumes. Braz J Med Biol Res. 1999;32(6):703-717. https://doi.org/10.1590/S0100-879X1999000600006
15. Pereira CA, Sato T, Rodrigues SC. New reference values for forced spirometry in white adults in Brazil. J Bras Pneumol. 2007;33(4):397-406. https://doi.org/10.1590/S1806-37132007000400008
16. Aidar M, Line SR. A simple and cost-effective protocol for DNA isolation from buccal epithelial cells. Braz Dent J. 2007;18(2):148-152. https://doi.org/10.1590/S0103-64402007000200012
17. Kim SK, Kang SW, Chung JH, Lee JS, Park HK, Yoon KL, et al. Coding single-nucleotide polymorphisms of interleukin-1 gene cluster are not associated with Kawasaki disease in the Korean population. Pediatr Cardiol. 2011;32(4):381-385. https://doi.org/10.1007/s00246-010-9858-7
18. Rad IA, Mohebbi I, Bagheri M. Molecular Evaluation of the IFN γ +874, TNF a -308, and IL-1Ra VNTR Sequences in Silicosis. Maedica (Bucur). 2012;7(1):20-24.
19. Wagner GR. Asbestosis and silicosis. Lancet. 1997;349(9061):1311-1315. https://doi.org/10.1016/S0140-6736(96)07336-9
20. Border WA, Noble NA. Transforming growth factor beta in tissue fibrosis. N Engl J Med. 1994;331(19):1286-1292. https://doi.org/10.1056/NEJM199411103311907
21. Wu F, Qu Y, Tang Y, Cao D, Sun P, Xia Z. Lack of association between cytokine gene polymorphisms and silicosis and pulmonary tuberculosis in Chinese iron miners. J Occup Health. 2008;50(6):445-454. https://doi.org/10.1539/joh.L8006
22. Grainger DJ, Heathcote K, Chiano M, Snieder H, Kemp PR, Metcalfe JC, et al. Genetic control of the circulating concentration of transforming growth factor type beta1. Hum Mol Genet. 1999;8(1):93-97. https://doi.org/10.1093/hmg/8.1.93
23. Barbarin V, Nihoul A, Misson P, Arras M, Delos M, Leclercq I, et al. The role of pro- and anti-inflammatory responses in silica-induced lung fibrosis. Respir Res. 2005;6(1):112. https://doi.org/10.1186/1465-9921-6-112
24. Jagirdar J, Begin R, Dufresne A, Goswami S, Lee TC, Rom WN. Transforming growth factor-beta (TGF-beta) in silicosis. Am J Respir Crit Care Med. 1996;154(4 Pt 1):1076-1081. https://doi.org/10.1164/ajrccm.154.4.8887610
25. Vodovotz Y, Bogdan C, Paik J, Xie QW, Nathan C. Mechanisms of suppression of macrophage nitric oxide release by transforming growth factor beta. J Exp Med. 1993;178(2):605-613. https://doi.org/10.1084/jem.178.2.605
26. Terrell TG, Working PK, Chow CP, Green JD. Pathology of recombinant human transforming growth factor-beta 1 in rats and rabbits. Int Rev Exp Pathol. 1993;34 Pt B:43-67. https://doi.org/10.1016/B978-0-12-364935-5.50009-2
27. Grigorova A, Trenova A, Stanilova S. A link between promoter polymorphisms of the transforming growth factor b1 (TGFB1) and TGF-β1 receptor II (TGFBR2) genes and relapsing-remitting multiple sclerosis. Folia Neuropathol. 2020;58(4):307-316. https://doi.org/10.5114/fn.2020.102433
28. Jin G, Deng Y, Miao R, Hu Z, Zhou Y, Tan Y, et al. TGFB1 and TGFBR2 functional polymorphisms and risk of esophageal squamous cell carcinoma: a case-control analysis in a Chinese population. J Cancer Res Clin Oncol. 2008;134(3):345-351. https://doi.org/10.1007/s00432-007-0290-1
29. Yao SQ, Rojanasakul LW, Chen ZY, Xu YJ, Bai YP, Chen G, et al. Fas/FasL pathway-mediated alveolar macrophage apoptosis involved in human silicosis. Apoptosis. 2011;16(12):1195-1204. https://doi.org/10.1007/s10495-011-0647-4
30. Suda T, Takahashi T, Golstein P, Nagata S. Molecular cloning and expression of the Fas ligand, a novel member of the tumor necrosis factor family. Cell. 1993;75(6):1169-1178. https://doi.org/10.1016/0092-8674(93)90326-L
31. Yu X, Li Y, Yu Y, Lei J, Wan G, Cao F. Associations between FAS rs2234767 and FASL rs763110 polymorphisms and the risk of lung cancer: a meta-analysis of 39,736 subjects. Onco Targets Ther. 2016;9:2049-2056. https://doi.org/10.2147/OTT.S102723
32. Borges VM, Falcão H, Leite-Júnior JH, Alvim L, Teixeira GP, Russo M, et al. Fas ligand triggers pulmonary silicosis. J Exp Med. 2001;194(2):155-164. https://doi.org/10.1084/jem.194.2.155
33. Huang P, Wang CH, Zhuo LY, Xia XS, Yang S, Zhang JW, et al. Polymorphisms rs763110 in FASL is linked to hepatitis C virus infection among high-risk populations. Br J Biomed Sci. 2020;77(3):112-117. https://doi.org/10.1080/09674845.2020.1747182
34. Wu S, Wang S, Fu Y, Tang W, Jin H, Meng Q, et al. A novel mechanism of rs763110 polymorphism contributing to cervical cancer risk by affecting the binding affinity of C/EBPβ and OCT1 complex to chromatin. Int J Cancer. 2017;140(4):756-763. https://doi.org/10.1002/ijc.30490
35. Khalifa RH, Bahgat DM, Darwish HA, Shahin RM. Significant association between FasL gene -844T/C polymorphism and risk to hepatocellular carcinoma in Egyptian patients. Immunol Lett. 2016;172:84-88. https://doi.org/10.1016/j.imlet.2016.02.007
36. Requena-Mullor M, Alarcón-Rodríguez R, Parrón-Carreño T, Martínez-López JJ, Lozano-Paniagua D, Hernández AF. Association between Crystalline Silica Dust Exposure and Silicosis Development in Artificial Stone Workers. Int J Environ Res Public Health. 2021;18(11):5625. https://doi.org/10.3390/ijerph18115625
37. Moreira VB, Ferreira A, Gabetto JM, Marchiori E, Lourenço PM. Comparative study of high resolution computer-assisted tomography with chest radiograph in silicosis [Article in Portuguese]. Rev Port Pneumol. 2003;9(1):33-40. https://doi.org/10.1016/S0873-2159(15)30658-9
38. Bégin R, Bergeron D, Samson L, Boctor M, Cantin A. CT assessment of silicosis in exposed workers. AJR Am J Roentgenol. 1987;148(3):509-514. https://doi.org/10.2214/ajr.148.3.509
39. Bégin R, Ostiguy G, Cantin A, Bergeron D. Lung function in silica-exposed workers. A relationship to disease severity assessed by CT scan. Chest. 1988;94(3):539-545. https://doi.org/10.1378/chest.94.3.539